Why Seeq API?¶
While the cost of next generation sequencing has continuously dropped in the last decade, data analysis and interpretation remains a challenging and expensive step in the precision medicine workflow in both clinical and research contexts. Seeq API solves two main problems in this space that remain unaddressed by existing tools.
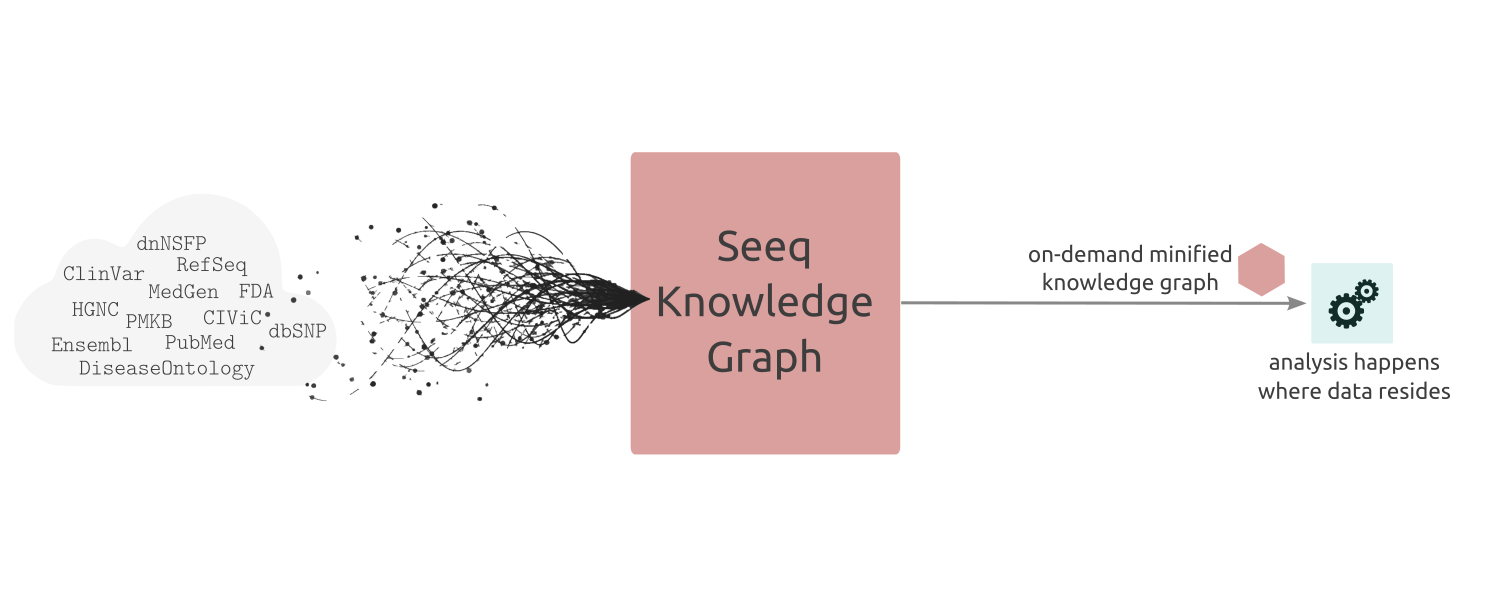
Knowledge Interoperability¶
There are dozens of valuable publicly available datasets like ClinVar, Cosmic, Ensembl, ChEMBL, and many others, each including a subset of the body of biological and clinical knowledge about the human genome. Unfortunately, these datasets often have mismatching data models, similar yet distinct semantics, and incompatible entity identification systems. Building scalable and traceable data pipelines for human genomics requires a mechanism for unifying all this knowledge under a simple, common vocabulary. Seeq API provides this interoperability layer across these knowledge silos.
Bring Algorithms to Data¶
Existing tools for variant analysis like SnpEff, Annovar, and VEP require either special-purpose compute environments, a technical bioinformatics support team, or both.
We found the status quo for genomic data analysis to be unnecessarily painful, slow, and error-prone. Let’s say a patient’s genome has been sequenced and you want to scan a VCF for variants of clinical significance. The typical workflow in a clinical or academic setting requires coordinating across different teams, waiting multiple hours or days, and shuffling sensitive PHI across multiple data centers.
The key insight of Seeq API is to build a knowledge graph that can slice and minify itself, on demand, for a particular analysis. This allows us to bring algorithms to data and in doing so enable clinicians, researchers, and bioinformaticians to perform ultra-fast reliable analyses at the network edge, on their consumer devices, and without any PHI leaving its origin.
Seeq VCF demonstrates the power of the slicing ability of the Seeq knowledge graph.