Seeq Knowledge Graph¶
The real power or Seeq API is in its knowledge graph built from thousands of publications and public datasets. The knowledge graph consists of multiple distinct graphs, each representing a specific semantic relationship between two or more core entities.
The most important feature of the knowledge graph is its ability to slice and minify itself for a particular analysis. This allows us to ship a small version of the knowledge graph to the machine where analysis happens. This means you can use Seeq API to perform your analyses where your data resides, without having to send your sensitive data to our servers.
Semantic atoms are n-ary relationships¶
In Seeq API n-ary relationships are first class citizens. They are the foundation of how the data is modeled and how the search API works. An n-ary relationship, as opposed to a binary relationship, is one that involves multiple entities of different types.
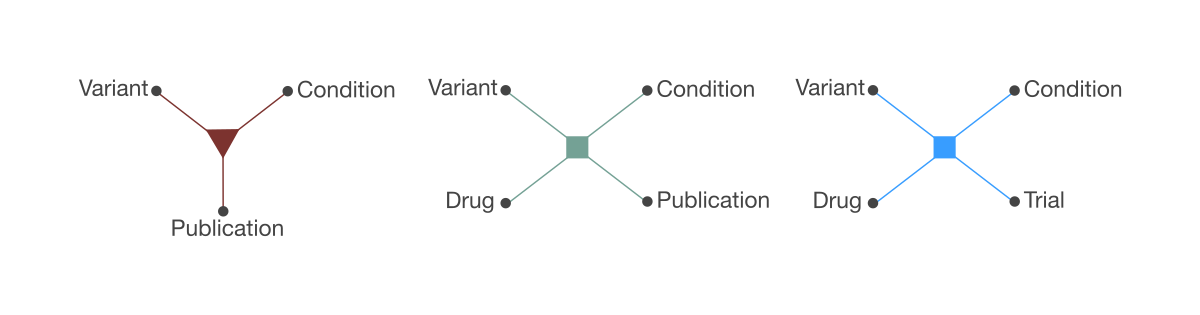
For example, one n-ary relationship could be concerned with evidence of pathogenicity of a certain variant in a certain disease. An instance of this relationship would correspond to a statement like:
Publication P supports pathogenicity of variant V of gene G in disease D.
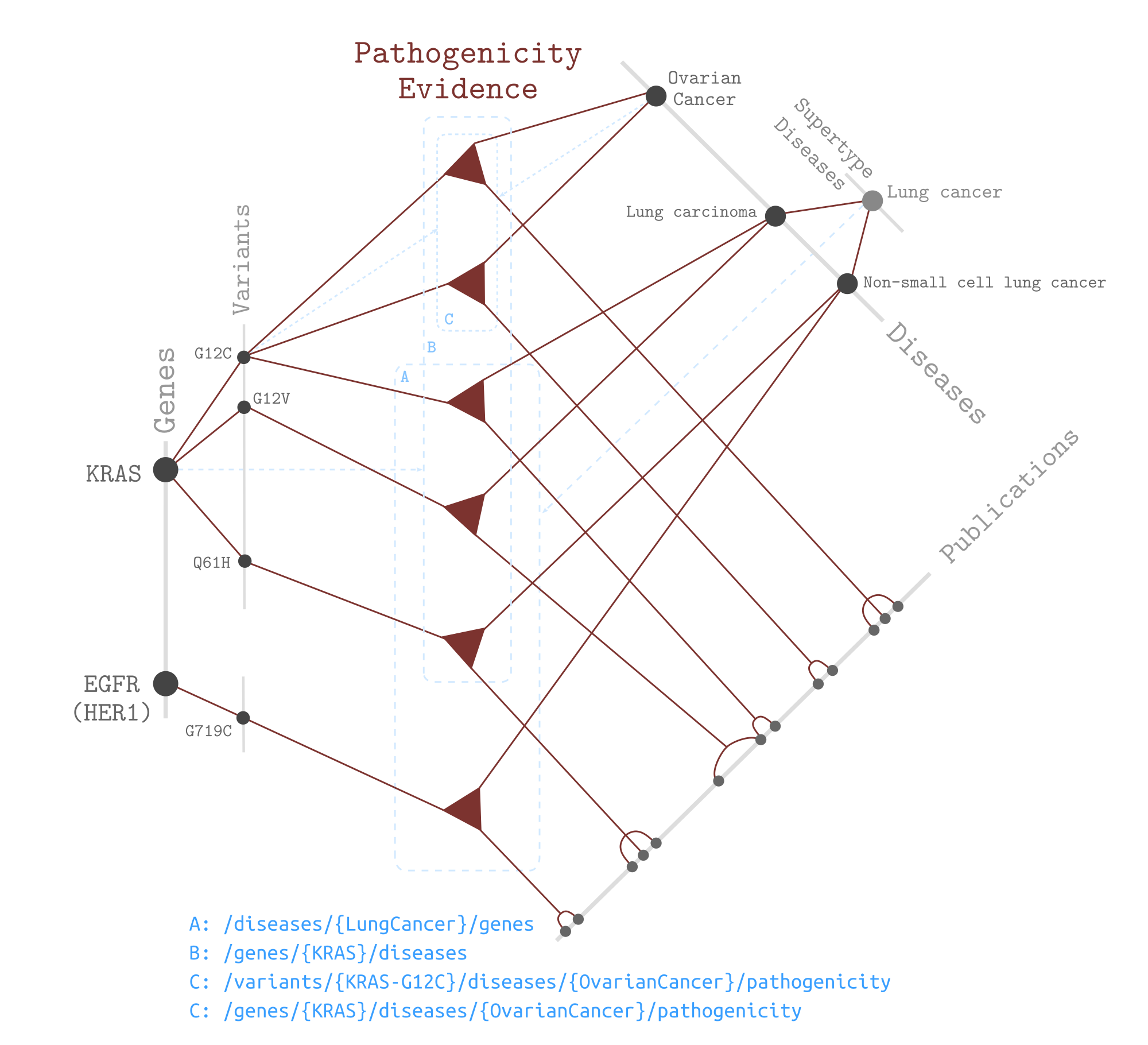
Pathogenicity Graph¶
The pathogenicity graph is concerned with evidence of pathogenicity connecting variants and diseases backed by a number of publications.
As an example graph to demonstrate the data model of Seeq’s n-ary relationships, the diagram below depicts the data model for the pathogenicity relationship, and how its instances are involved in various search scenarios.
Treatability Graph¶
The treatability graph is composed of multiple distinct graphs itself:
Targeted treatments for a variant in a certain disease: the entities involved in this graph are genes, variants, diseases, drugs, and publications.
Gene-drug interactions: the entities involved in this graph are genes, drugs, and publications. Assertions in this graph do not have a specific disease context nor do they involve a specific variant.
Drug-disease indications: the entities involved in this graph are drugs, diseases, and publications. Assertions in this graph do no have a specific drug or variant context.
Graph Search¶
Seeq’s Graph search API includes two steps: search for connected entities, and extract supported evidence for each connection.
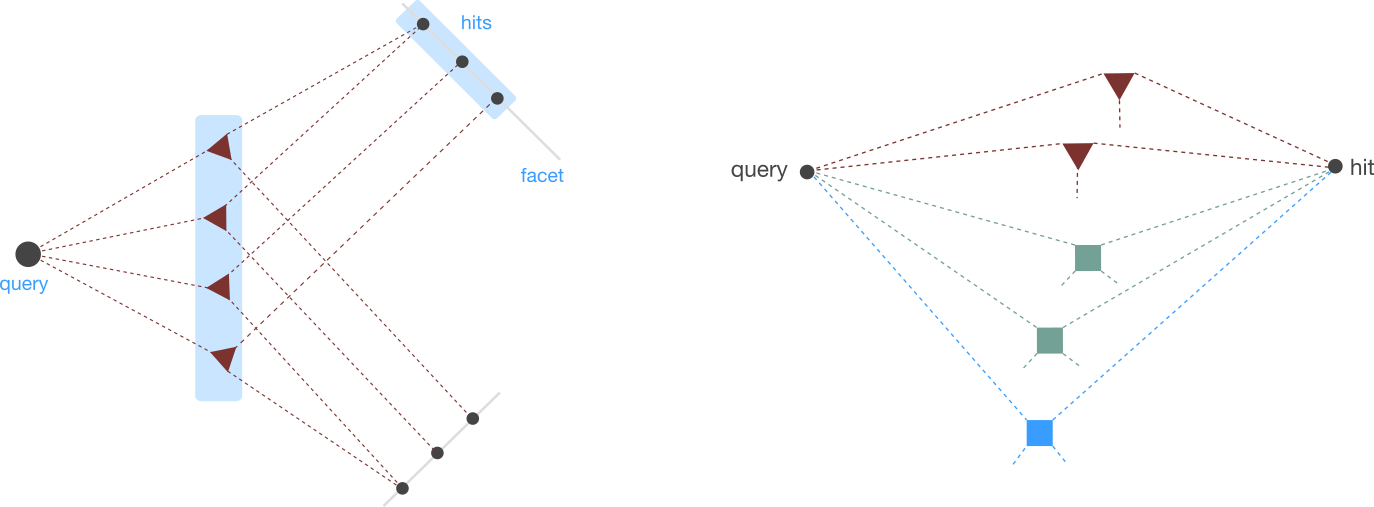
Connected Entities¶
The first set of search endpoints are concerned with the following problem. Given a specific query entity Q (say some gene), find all entities of type T (say diseases) that are connected to Q through either of the n-ary relationships.
For example, let’s say we want to find out all diseases that are connected to gene IDH1, ranked by number and strength of the available evidence:
$ curl https://api.seeq.bio/genes/3417/diseases
Note
This endpoint corresponds to the search results you get on this page of seeq.bio.
The first hit is Acute myeloid leukemia (AML):
[
{
"hit": { "cui": "C0023467", "name": "Acute myeloid leukemia" },
"treatability": {
"disease": "...",
"summary": {
"max_clinsig": "positive",
"evidence_count": 4,
"text": "4 evidence records on targeted treatments in Acute myeloid leukemia with IDH1 mutations, some show positive response."
}
},
"pathogenicity": {
"disease": "...",
"summary": {
"max_clinsig": "pathogenic",
"evidence_count": 6,
"text": "6 evidence records for IDH1 variants in Acute myeloid leukemia, some are pathogenic."
}
}
},
"..."
]
Similar endpoints are available for all pairs of entities, see the API reference for more details.
Supporting Evidence¶
The second set of endpoints are concerned with the following problem. Given a specific query entity Q and a specific search hit entity H from the previous step, find all instances of all n-ary relationships that support that connection.
In the example above, let’s say we want to find the supporting evidence for the connection between the gene IDH1 and the disease AML. To do this, we need to query each of the implicated subgraphs separately. The pathogenicity evidence for the connection is backed by the pathogenicity graph:
$ curl https://api.seeq.bio/genes/3417/diseases/C0023467/pathogenicity
Each entry in the response is a single instance of the n-ary pathogenicity relationship and counts as a unit of evidence for the connection between the gene and disease of interest:
[
{
"gene": { "entrez_id": "3417", "canonical_symbol": "IDH1", "..." },
"disease": { "cui": "C0023467", "name": "Acute myeloid leukemia" },
"variant": "R132L",
"clinsig": "pathogenic"
"citation": {
"source": "ClinVar",
"variation_id": "375889",
"pmids": [ 22160010, 22397365, 22417203, 22898539, 26619011 ],
"publications": [
{
"pmid": 26619011,
"title": "Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity.",
"journal": "Nat Biotechnol",
"year": 2016,
"first_author": "Chang"
},
"...",
],
},
},
"...",
]
And the other set of evidence is backed by the treatability graph:
$ curl https://api.seeq.bio/genes/3417/diseases/C0023467/treatability
Each entry in the response is a now single instance of the n-ary treatability relationship:
[
{
"gene": { "entrez_id": "3417", "canonical_symbol": "IDH1", "..." },
"disease": { "cui": "C0023467", "name": "Acute myeloid leukemia" },
"variant": null,
"drug": {
"chembl_id": "CHEMBL3989958",
"name": "Ivosidenib",
"synonyms": [],
"fda_approved": true,
"development_phase": 4
},
"clinsig": "positive",
"source": "CIViC",
"citation": {
"citation_type": "PubMed",
"citation_ids": [ "29860938" ]
"text": "I phase 1 trial for patients with IDH1-mutated AML, patients who received ivosidenib (AG-120). The rate of complete remission was 21.6%, the over all response rate was 30.4%.",
"publications": [
{
"pmid": 29860938,
"title": "Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML.",
"journal": "N Engl J Med",
"year": 2018,
"first_author": "DiNardo"
}
]
}
},
"..."
]
MicroSeeq¶
Seeq’s knowledge graph is designed so that it can be sliced and minified, on demand, for each particular analysis context. This minified knowledge graph is called MicroSeeq.
MicroSeeq enables the Seeq client-side SDK to build complex variant interpretation pipelines like Seeq VCF that run entirely on the client side (e.g. your browser, or your virtual machine) without your raw variant PHI leaving your device.
To obtain a MicroSeeq, all you need to do is specify your genes of interest:
$ curl https://api.seeq.bio/micro-seeq?ids=3417 # genes of interest: IDH1
{
"skeletons": [
{
"gene": {
"entrez_id": "3417",
"canonical_symbol": "IDH1",
"..."
},
"exons": [
{
"ense": "ENSE00001801507",
"enst": "ENST00000415913",
"rank": 1,
"chrom_start": 208254031,
"chrom_end": 208254322,
"fn_cdna_pos": 1,
"ln_cdna_pos": 292,
"..."
},
"..."
],
"transcript": {
"ensg": "ENSG00000138413",
"enst": "ENST00000415913",
"ensp": "ENSP00000390265",
"nm": null,
"chrom": "2",
"n_exons": 10,
"fcn_cdna_pos": 383,
"lcn_cdna_pos": 1627,
"cds_length": 1245,
"..."
"cdna": "AGGGGAG...",
"peptide": "MSKKISA..."
},
"domains": [
{
"pfam_id": "PF00180",
"pfam_name": "Iso_dh",
"name": "Isocitrate/isopropylmalate dehydrogenase",
"summary": null,
"aa_start": 11,
"aa_end": 399
}
]
}
],
"evidence": [
{
"gene": {
"entrez_id": "3417",
"canonical_symbol": "IDH1",
"..."
},
"pathogenicity": [
{
"eal_cds_pos": 394,
"max_clinsig_rank": 10
},
{
"eal_cds_pos": 395,
"max_clinsig_rank": 10
}
],
"treatability": [
{
"eal_cds_pos": 394,
"max_clinsig_rank": 80
},
{
"eal_cds_pos": 395,
"max_clinsig_rank": 50
}
],
"gene_treatability": 80
}
],
"..."
}